BioKC
A collaborative platform for systems biology model curation and annotation
These are the documentation pages of BioKC platform, a web-based collaborative platform for the curation and annotation of biomedical knowledge following the standard data model from Systems Biology Markup Language (SBML).
This novel platform enables the user to construct building blocks of systems biology models and allows the user to annotate them with human-provided and machine-identified literature evidence. Knowledge representation in BioKC follows the SBML standard in formalising elements of a given model, their relationships and annotations.
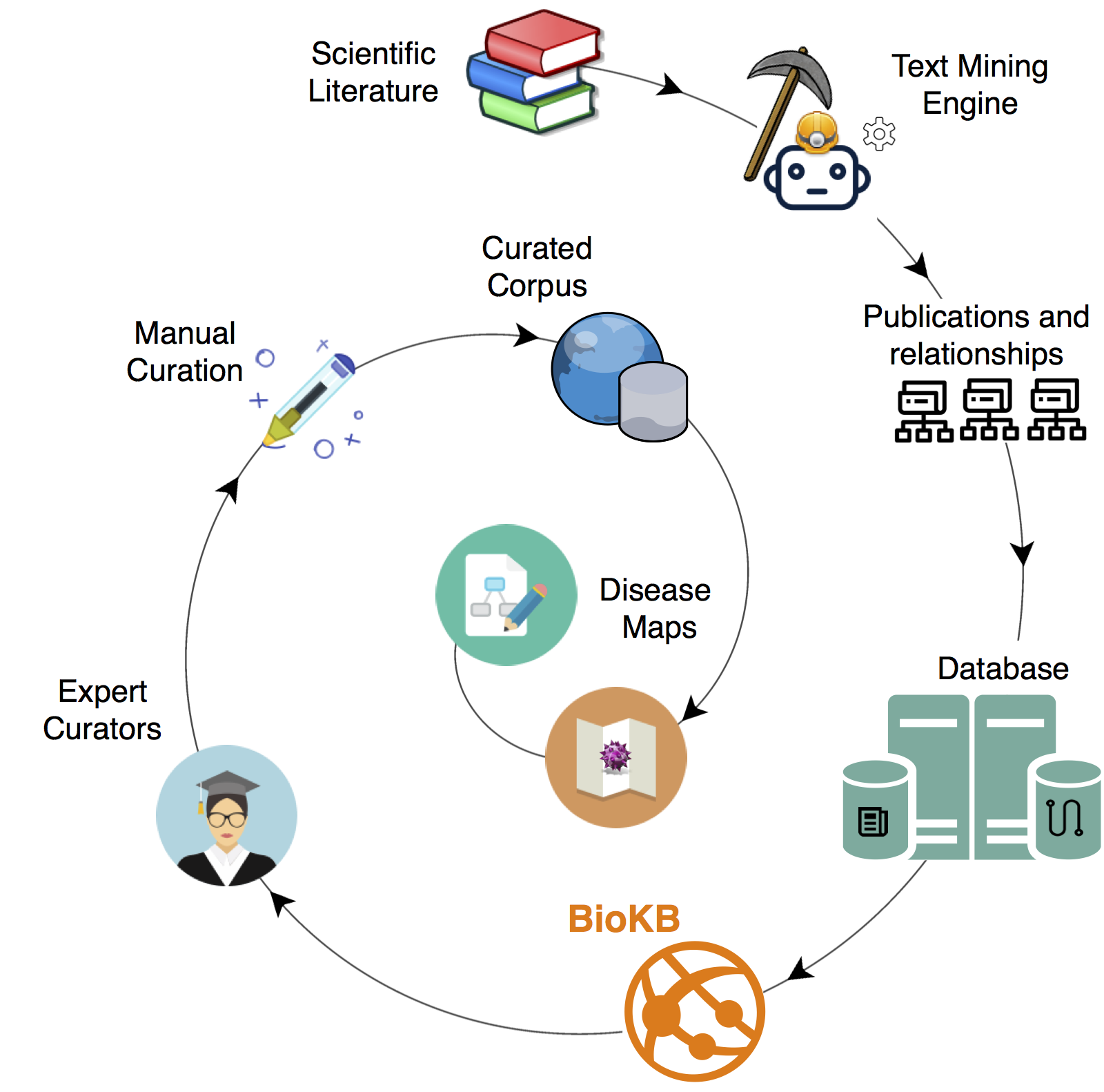
BioKB platform#
BioKC is built on top of BioKB, a web-based interface designed to browse the text-mining results of almost 30 million publications including both abstracts and full-text articles.
Try it at: https://biokb.lcsb.uni.lu/
Cite as ✍️#
Our latest publication is available at BioRxiv. Nevertheless, we are currently working on a full journal manuscript based on this publication.
Vega, Carlos; Grouès, Valentin; Ostaszewski, Marek; Schneider, Reinhard; & Satagopam, Venkata. (2020). BioKC: a collaborative platform for systems biology model curation and annotation. BioRxiv. https://doi.org/10.1101/2020.10.01.322438
Other publications#
We had the pleasure to participate on the last European Conference on Computational Biology (ECCB2020) during the workshop BioNetVisA. Poster, slides, and video presentation can be found at Zenodo.
Vega, Carlos; Grouès, Valentin; Ostaszewski, Marek; Satagopam, Venkata; & Schneider, Reinhard. (2020). BioKC: a platform for quality controlled curation and annotation of systems biology models. Zenodo. http://doi.org/10.5281/zenodo.4033071
To cite BioKB platform#
BioKB website is the front-end of BioKB platform, a pipeline which, by exploiting text mining and semantic technologies, helps researchers easily access semantic content of thousands of abstracts and full text articles. The text mining component analyzes the articles content and extracts relations between a wide variety of concepts, extending the scope from proteins, chemicals and pathologies to biological processes and molecular functions. Extracted knowledge is stored in a knowledge base publicly available for both, human and machine access, via this web application and SPARQL endpoint.
Biryukov, M., Groues, V., Satagopam, V.; & Schneider, R. (2018).BioKB-Text Mining and Semantic Technologies for Biomedical Content Discovery. Figshare. https://doi.org/10.6084/m9.figshare.6994121
Contributors 👨🏼💻👩🏽💻👨🏻💻👩🏼💻👨🏿💻#
This is a project of the Bioinformatics Core research group from the Luxembourg Centre for Systems Biomedicine, in the University of Luxembourg.
The main contributors to the project are Carlos Vega, Valentin Grouès, Marek Ostaszewski, Venkata Satagopam and Reinhard Schneider.
We would like to acknowledge Valentin Devassine for his work with end to end testing and Christos Kyriazis for his contributions to the unit tests.
Acknowledgements#
The authors would like to thank the Luxembourg National Research Fund (FNR) for supporting this work through grant 14729738 for Covid19 Literature Bio-curation, Text-mining And Semantic Web Technologies (COVlit) and the National Centre of Excellence in Research on Parkinson’s disease (NCER-PD [FNR/NCER13/BM/11264123])